Reproducibility in Cancer Biology: Challenges for assessing replicability in preclinical cancer biology
Figures
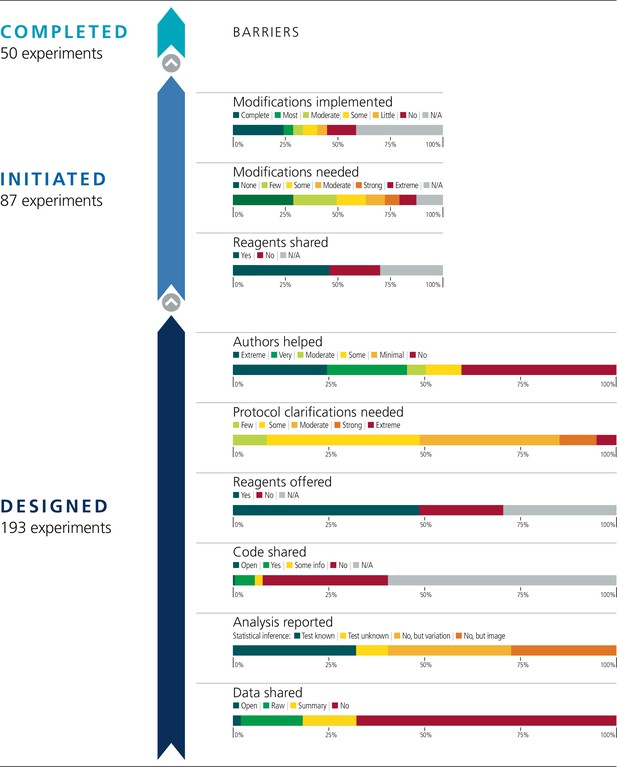
Barriers to conducting replications – by experiment.
During the design phase of the project the 193 experiments selected for replication were coded according to six criteria: availability and sharing of data; reporting of statistical analysis (i.e., did the paper describe the tests used in statistical analysis?; if such tests were not used, did the paper report on biological variation (e.g., graph reporting error bars) or representative images?); availability and sharing of analytic code; did the original authors offer to share key reagents?; what level of protocol clarifications were needed from the original authors?; how helpful were the responses to those requests? The 29 Registered Reports published by the project included protocols for 87 experiments, and these experiments were coded according to three criteria: were reagents shared by the original authors?; did the replication authors have to make modifications to the protocol?; were these modifications implemented? A total of 50 experiments were completed.
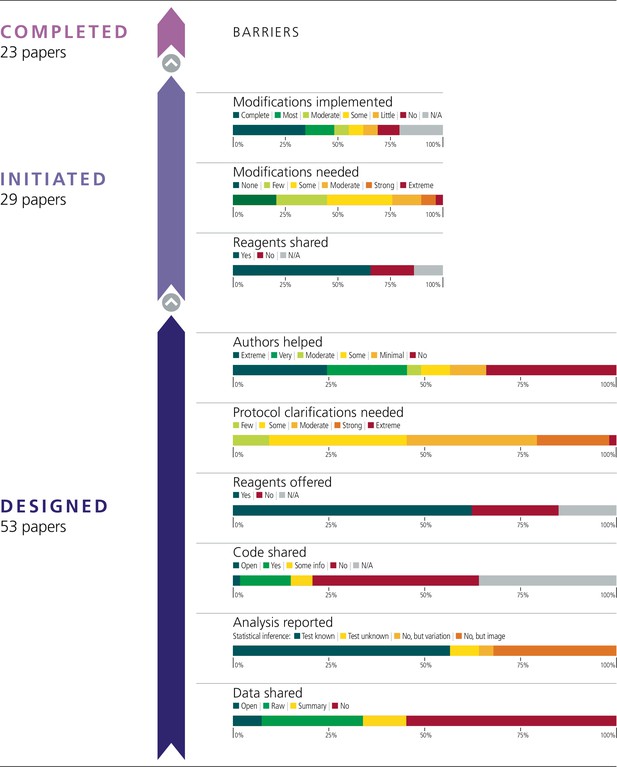
Barriers to conducting replications – by paper.
This figure is similar to Figure 1, except that the results for the experiments have been combined to give results for the papers that contained the experiments. Experiments from 53 papers were coded during the design phase; experiments from 29 papers were initiated; and results from 23 papers were published. Two methods were used to convert the scores for experiments into scores for papers. For four criteria (protocol clarifications needed; authors helped; modifications needed; modifications implemented) the average of the scores from the experiments was assigned to the paper. For five criteria (data shared; analysis reported; code shared; reagents offered; reagents shared), scoring was conducted with a 'liberal' interpretation for sharing: for example, if data were shared for just one experiment in a paper, the paper was coded as data shared. See main text for further details.
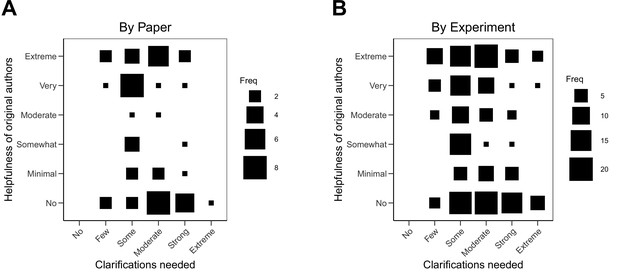
Relationship between extent of clarification needed and helpfulness of authors.
Fluctuation plots showing the coded ratings for extent of clarifications needed from original authors and the degree to which authors were helpful in providing feedback and materials for designing the replication experiments. The size of the square shows the number (Freq) of papers/experiments for each combination of extent of clarification needed and helpfulness. (A) To characterize papers (N = 53), coded ratings were averaged across experiments for each paper. The average number of experiments per paper was 3.6 (SD = 1.9; range = 1–11). The Spearman rank-order correlation between extent of clarification needed and helpfulness was –0.24 (95% CI [–0.48, 0.03]) across papers. (B) For experiments (N = 193), the Spearman rank-order correlation between extent of clarification needed and helpfulness was –0.20 (95% CI [–0.33, –0.06]).

Techniques used in the original experiments.
A total of 820 experimental techniques were identified in the 193 experiments selected for replication at the start of the project. These techniques were coded into 48 sub-categories, which were grouped into four categories (cell assays; immunoassays; animal assays; molecular assays).
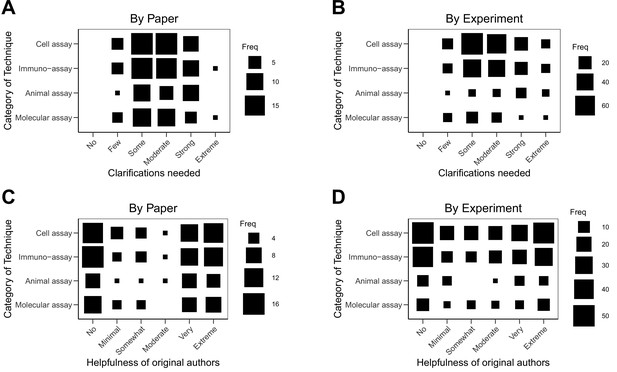
Relationship between extent of clarification needed or helpfulness with category of techniques.
Fluctuation plots showing the coded ratings for the category of techniques used in the original experiments and the extent of clarification needed from the original authors by paper (A) and by experiment (B). Fluctuation plots showing the category of techniques and the helpfulness of the original authors by paper (C) and by experiment (D). The size of the square shows the number (Freq) of papers/experiments for each combination. The average number of categories used in the 193 experiments was 2.2 (SD = 0.7; range = 1–4). To characterize papers (N = 53), coded ratings were averaged across the experiments for each paper; the average number of categories used per paper was 2.9 (SD = 0.7; range = 1–4).
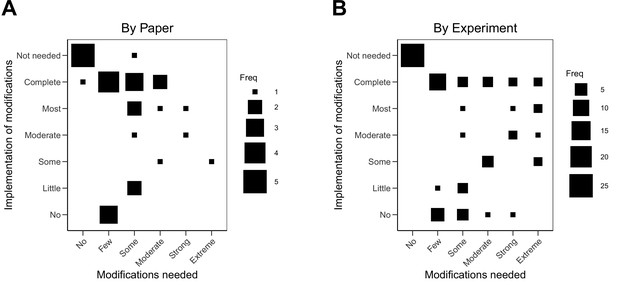
Relationship between extent of modifications needed and implementation of modifications.
Fluctuation plots showing the coded ratings for extent of modifications needed in order to conduct the replication experiments, and the extent to which the replication authors were able to implement these modifications for experiments that were conducted. The size of the square shows the number (Freq) of papers/experiments for each combination. (A) To characterize papers (N = 29), coded ratings were averaged across the experiments conducted for each paper. The average number of experiments conducted per paper was 2.6 (SD = 1.3; range = 1–6), and the Spearman rank-order correlation between extent of modifications needed and implementation was –0.01 (95% CI [–0.42, 0.40]). (B) For the experiments that were started (N = 76), the Spearman rank-order correlation was 0.01 (95% CI [–0.27, 0.28]).
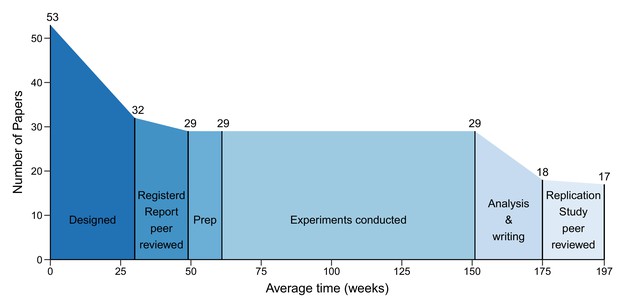
The different phases of the replication process.
Graph showing the number of papers entering each of the six phases of the replication process, and the mean duration of each phase in weeks. 53 papers entered the design phase, which started with the selection of papers for replication and ended with submission of a Registered Report (mean = 30 weeks; median = 31; IQR = 21–37). 32 papers entered the protocol peer reviewed phase, which ended with the acceptance of a Registered Report (mean = 19 weeks; median = 18; IQR = 15–24). 29 papers entered the preparation phase (Prep), which ended when experimental work began (mean = 12 weeks; median = 3; IQR = 0–11). The mean for the prep phase was much higher than the median (and outside the IQR) because this phase took less than a week for many studies, but much longer for a small number of studies. The same 29 papers entered the conducted phase, which ended when the final experimental data were delivered (mean = 90 weeks; median = 88; IQR = 44–127), and the analysis and writing phase started, which ended with the submission of a Replication Study (mean = 24 weeks; median = 23; IQR = 7–32). 18 papers entered the results peer review phase, which ended with the acceptance of a Replication Study (mean = 22 weeks; median = 18; IQR = 15–26). In the end, 17 Replication Studies were accepted for publication. The entire process had a mean length of 197 weeks and a median length of 181 weeks (IQR = 102–257).
Tables
The 53 papers selected for replication in the RP:CB.
-
193 experiments in 53 papers were selected for replication. The papers are listed in column 1, and the number of experiments selected from each paper is listed in column 2. Registered Reports for 87 experiments from 29 papers were published in eLife. The Registered Reports are listed in column 3, and the number of experiments included in each Registered Report is listed in column 4. 50 experiments from 23 Registered Reports were completed. 17 Replication Studies reporting the results of 41 experiments were published in eLife; the results of another nine experiments from the six remaining Registered Reports were published in an aggregate paper (Errington et al., 2021a). The Replication Studies are listed in column 5, and the number of experiments included in each study is listed in column 6. Column seven contains a link to data, digital materials, and code.





